Our Research
Science drives everything we do. Here, you’ll find PURR.AI’s peer-reviewed publications, conference papers, and research collaborations. Each reflects our commitment to advancing AI-driven drug discovery and sharing knowledge with the global scientific community.
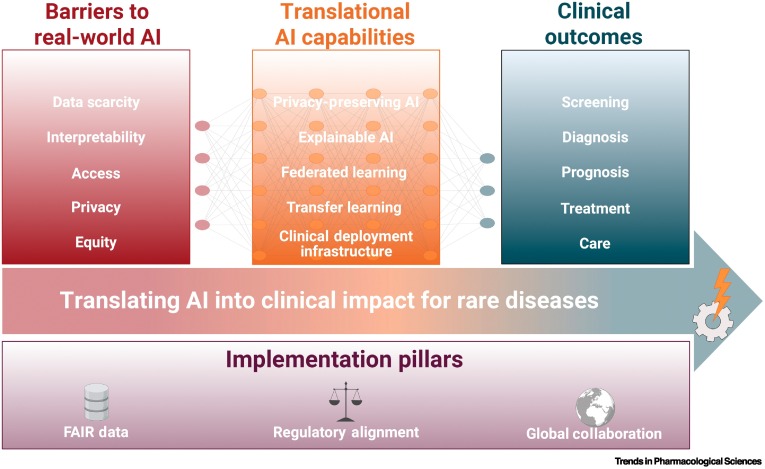
This review shows how cutting-edge artificial intelligence can transform rare disease care by accelerating diagnosis, guiding personalised treatment, and enabling faster, more efficient drug discovery.
It highlights that real clinical impact depends not only on powerful algorithms, but on high-quality FAIR data, privacy-preserving collaboration, explainable models, and globally coordinated, equity-focused infrastructures.
We systematically examined recent advances in AI for rare diseases across screening, diagnosis, prognosis, and therapy development, spanning methods such as transfer learning, generative models, federated learning, NLP, and explainable AI.
We then mapped the technical, ethical, and infrastructural priorities needed to move from proof-of-concept tools to trustworthy, integrated clinical solutions worldwide.
Despite clear promise, most AI tools for rare diseases remain fragmented and fail to reach real patients, largely due to data scarcity, bias, privacy constraints, and limited interoperability between healthcare systems.
By distilling best practices and common pitfalls, this work provides a roadmap to align innovators, clinicians, regulators, and patient communities around responsible, scalable AI adoption.
The review validates PURR.AI’s strategy of building FAIR-by-design, privacy-preserving, explainable AI platforms that integrate genomics, clinical data, and molecular knowledge for rare diseases.
It positions PURR.AI to partner as an infrastructure and analytics provider that can turn complex multi-omic data into actionable, equitable clinical insights.
GPCR‑A17 MAAP is a machine‑learning tool that predicts whether small molecules act as agonists, antagonists, or modulators across the GPCR‑A17 receptor subfamily.
It streamlines early drug discovery by replacing years of screening with rapid, high‑accuracy predictions.
We built an ensemble AI model (XGBoost, Random Forest, LightGBM) trained on 6,900 curated receptor‑ligand pairs, combining chemical descriptors with protein sequence embeddings.
The model predicts ligand function across 21 GPCR‑A17 receptors with state‑of‑the‑art accuracy (up to 0.93 F1 on Ki‑filtered datasets).
Determining how compounds interact with GPCRs has been slow, costly, and uncertain. Our model accelerates this process, enabling rational design, repurposing, and prioritisation of safer, more effective drug candidates.
GPCR‑A17 MAAP strengthens PURR.AI’s AI‑drug discovery platform by:
Reducing R&D timelines through fast, data‑driven target mapping.
Expanding discovery reach to receptors often avoided due to cost and complexity.
Improving portfolio value with more precise lead prioritisation and de‑risking of development pipelines.
This review highlights recent advances in computational and AI-driven methods for predicting drug toxicity, emphasizing the importance of drug-target binding affinity (DTBA) in anticipating adverse effects.
It surveys state-of-the-art models, key toxicity endpoints, and emerging machine learning strategies that can accelerate safer drug development and reduce reliance on animal testing.
We systematically reviewed the current landscape of computational approaches, especially artificial intelligence (AI) and machine learning (ML) models, for toxicity prediction in drug development.
This included a critical evaluation of breakthrough algorithms for major endpoints (e.g., LD50, DILI, hERG, carcinogenicity, Ames mutagenicity), as well as models for DTBA prediction—a key molecular feature in toxicity and efficacy assessment.
We compared data sources, modeling strategies, and performance metrics, providing the community with a comprehensive resource and benchmarking of current methods.
Late-stage drug attrition remains high, with unexpected toxicity being a major contributor to costly failures. Existing experimental toxicity testing is resource-intensive, time-consuming, and ethically challenging due to animal use.
By consolidating the latest computational advances—particularly in AI/ML-based toxicity and DTBA prediction—we aim to promote more accurate, rapid, and humane methods to screen drug candidates early in development. This addresses regulatory needs for safety while accelerating translation from discovery to clinic.
For PURR.AI, leveraging advanced AI methodologies in toxicity and DTBA prediction aligns perfectly with our mission to deliver smarter, more predictive molecular modeling solutions.
The approaches summarized in this review provide a scientific foundation for enhancing our platform’s safety profiling modules, reducing the risk of late-stage failures for our clients, and supporting FAIR, 3Rs-compliant research.
Integrating these state-of-the-art computational strategies positions PURR.AI at the forefront of data-driven drug discovery and predictive toxicology.